Dysregulated thyroid hormone level is associated with a wide variety of metabolic complications. For example, hypothyroidism is associated with decreased basal metabolic rate, weight gain, increased serum cholesterol, non-alcoholic liver disease and type II diabetes. The molecular action of thyroid hormone is primarily mediated by the thyroid hormone receptors (TRs) acting as transcription factors to regulate expression of genes in major tissues such as the heart, adipose tissue, muscle and liver. A molecular bimodal switch model has for years been used to explain how TRs regulate gene expression. However, the model has not until now been rigorously tested on a genome-wide scale.
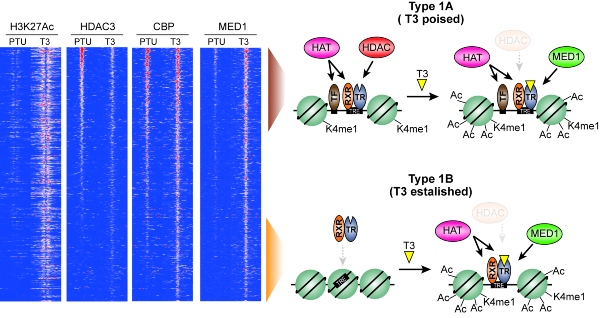
Figure 1. Revised model describing activation (histone hyperacetylation) of gene regulatory regions by thyroid hormone (T3). PTU represents hypothyroid animals.
In collaboration with researchers at NIH and Weill Cornell Medicine in the US, PhD student Stine Præstholm in the Grøntved group used genetically modified mice and next generation sequencing to test the current model. They successfully identified multiple different mechanisms controlling gene expression by T3. This includes histone hyperacetylation by loss of histone deacetylases (HDAC3) at some gene regulatory regions (defined as Type 1A regions) as well as recruitment of histone acetyltransferases (CBP) to other regions (defined as Type 1B regions). These new findings suggest a revision of the prevailing bimodal switch model. The researchers hope that this study, together with future studies, will add mechanistic knowledge to the molecular action of T3 that will benefit treatment of disorders associated with dysregulated T3 signaling.
The paper is published in PLOS Genetics and can be read in full here.
Correspondence may be directed to Lars Grøntved: larsgr@bmb.sdu.dk