Now researchers can follow the hectic life inside a cell
Living cells are constantly on the move. They move around and divide, and they are responsible for transporting molecules around inside themselves. Now SDU researchers have developed a method that makes it possible to become a spectator at this hectic traffic. The method is of particular importance for disease research.
Every cell in our body is constantly active. Cells are inhabited by a myriad of different molecules tirelessly interacting with each other to keep the machinery - your body – going.
In the course of one minute, almost all molecules within a cell have moved around to perform various tasks in the cell.
- There is a lot of activity and a lot of traffic inside a cell. And that's actually one of science's great mysteries: How can these constant molecular transport processes be performed with such precision and coordination, researchers Achim Schroll and Daniel Wüstner from University of Southern Denmark ask.
Together with PhD student Christian V. Hansen from the Department of Mathematics and Computer Science, they have developed a new model that makes it possible to monitor the lively traffic inside a single cell. Their work is published in the journal Computing and Visualization in Science.
Wrong movement can be fatal
The model is important, because sometimes the molecular transport processes are not performed properly, which may cause diseases.
- The consequences can be fatal. Many diseases develop because the transport is disturbed or because proteins clump in the cell. This is the case in diseases like Alzheimer's, Parkinson's and Huntington's. Therefore it is important to study molecules activities in cells, says Daniel Wüstner.
The researchers started their work by studying living cells under a microscope and observe how factors such as temperature and biochemical reactions make molecules move inside a cell. These observations were "translated" into a mathematical model based on differential equations.
- Thus we now have a computer model that allows us to run a simulation of what goes on inside a living cell. We know the terms "in vivo" and "in vitro" (in a living organism and in the test tube) - here we examine things "in silico"; in the computer, explains Achim Scholl.
What exactly did the researchers do?
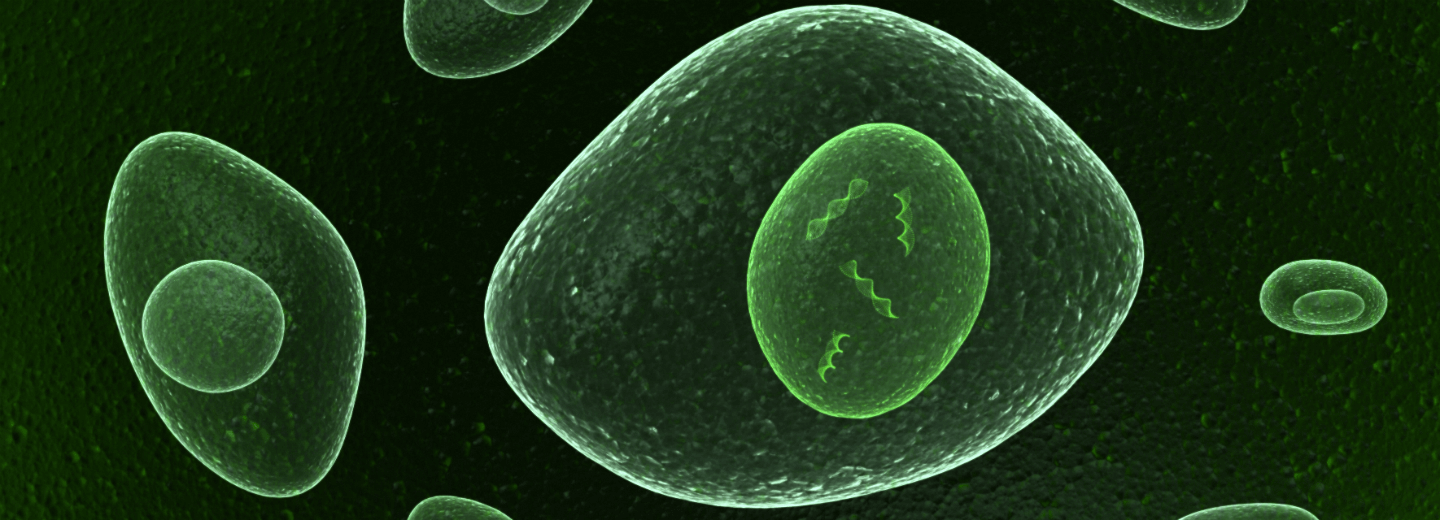
Molecules can be made to light up green, so you can see them in a microscope. They will appear as a green mass indicating that you that you are looking into a cell that is full of green molecules.
For more details the researchers installed an on/off-switch inside the cell. Each time a moving molecule passed through the switch, its green color got switched off. Gradually, more and more molecules in the cell became dark - a sign that more and more molecules had passed through the switch.
This traffic was "translated" into a mathematical model, so researchers can now use a computer to study the molecular traffic in a cell and see what happens if conditions change. One example could be that a membrane becomes harder for the molecules to penetrate, so it takes longer for them for them to reach the switch.
In real time, it takes approximately one minute for the majority of molecules to pass through the switch. After approximately 10 minutes all have passed through, and there are no more green lights to see.
Most molecules are quick to pass through, while certain conditions slow down the last ones. One reason is that some molecules are stronger bound to a particular structure. Another reason could be that they have to pass a barrier.
Paper: Computational modeling of fluorescence loss in photobleaching. Christian V. Hansen, Hans J. Schroll, Daniel Wüstner. Computing and Visualization in Science, Volume 17, Issue 4, pp 151-166.